load library and datasets
library(DOSE)
library(tidyverse)
library(org.Hs.eg.db)
library(clusterProfiler)
library(DT)
library(msigdbr)
get ranks of genes
# there are 5 comparisons. This dataset is from "Dietrich C, Trub A. INX-315, a selective CDK2 inhibitor, induces cell cycle arrest and senescence in solid tumors. Cancer Discov. 2023 Dec 4"
DE_list %>% names
## [1] "PAR_LYvsDMSO" "PAR_ARCvsDMSO" "PAR_LY.ARCvsDMSO" "LYR_ARCvsDMSO"
## [5] "LYFR_ARCvsDMSO"
## $PAR_LYvsDMSO
## A1BG-AS1 A4GALT AAAS AACS AAGAB AAK1
## -0.9150194 0.2292568 -0.5303841 -0.2728154 -0.2673752 1.1438151
##
## $PAR_ARCvsDMSO
## A1BG-AS1 A4GALT AAAS AACS AAGAB AAK1
## -0.33311195 0.12477299 -0.09373685 -0.11442974 -0.18108505 0.44105658
##
## $PAR_LY.ARCvsDMSO
## A1BG-AS1 A4GALT AAAS AACS AAGAB AAK1
## -0.57935270 0.12057293 -1.02542265 -0.02503685 0.03875984 0.84889094
##
## $LYR_ARCvsDMSO
## A1BG-AS1 A4GALT AAAS AACS AAGAB AAK1
## -0.1221634 0.2373029 -0.0791930 -0.1793278 -0.5279337 0.9647828
##
## $LYFR_ARCvsDMSO
## A1BG-AS1 A4GALT AAAS AACS AAGAB AAK1
## 0.1851126 0.4573528 -0.3331115 0.2801182 -0.1870958 1.2414269
# %>% filter(padj < 0.2) %>% arrange(desc(NES))
gsea_df$PAR_LYvsDMSO %>% datatable(caption = "PAR_LYvsDMSO")
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HALLMARK_E2F_TARGETS | HALLMARK_E2F_TARGETS | 200 | -0.792833534795824 | -3.36609513059318 | 2.17643566148187e-54 | 1.08821783074093e-52 | 4.35287132296373e-53 | 1848 | tags=68%, list=13%, signal=61% | LYAR/XRCC6/MSH2/SHMT1/MLH1/TP53/SUV39H1/LUC7L3/CDKN1B/TFRC/PHF5A/SMC6/NUDT21/POLD2/CTPS1/MRE11/PAICS/RBBP7/SLBP/PLK4/RAD51C/DONSON/UBR7/DCLRE1B/CDK4/STAG1/RANBP1/SNRPB/TRA2B/TUBG1/GINS3/CKS2/EXOSC8/SMC3/UNG/TIPIN/POLD3/TUBB/RPA3/UBE2T/PRKDC/POLD1/CCP110/SRSF1/SMC1A/RFC2/KPNA2/LBR/CKS1B/TRIP13/RACGAP1/H2AZ1/CDC25B/NAP1L1/HNRNPD/PSMC3IP/MCM4/DUT/BRCA2/NCAPD2/DNMT1/POLE/SSRP1/SRSF2/MCM3/USP1/UBE2S/ANP32E/MMS22L/MELK/DEK/NASP/KIF22/SMC4/MAD2L1/EZH2/TIMELESS/PCNA/LIG1/STMN1/MCM5/POLA2/CDCA8/MCM2/AURKA/DDX39A/CIT/ASF1B/GINS1/E2F8/TACC3/CCNB2/SPAG5/H2AX/MCM6/CENPE/ATAD2/TMPO/HELLS/CDC25A/BARD1/PSIP1/RAD51AP1/MCM7/SPC24/CBX5/HMMR/BRCA1/CDK1/CHEK1/CDKN3/TK1/CENPM/TCF19/DSCC1/DLGAP5/CDKN2C/RRM2/ESPL1/DEPDC1/CDC20/BIRC5/BUB1B/KIF4A/CDCA3/KIF18B/MKI67/KIF2C/PLK1/TOP2A/LMNB1/HMGB2/PTTG1/MYBL2/MXD3/AURKB/SPC25 |
| 2 | HALLMARK_INTERFERON_GAMMA_RESPONSE | HALLMARK_INTERFERON_GAMMA_RESPONSE | 159 | 0.852640357539166 | 2.90307464945179 | 1.34411785999181e-45 | 3.36029464997952e-44 | 1.34411785999181e-44 | 1055 | tags=58%, list=7%, signal=54% | RSAD2/OAS2/IFIT3/IFIT2/MX2/IFI27/IFIT1/XAF1/CMPK2/OASL/IFI44L/BATF2/DDX60/RTP4/TNFAIP2/DDX58/ISG15/IFI44/DHX58/TNFSF10/IFIH1/CCL5/APOL6/OAS3/BST2/UBE2L6/ISG20/PSMB9/PARP12/PSMB8/GBP4/USP18/LGALS3BP/PARP14/IRF9/TAP1/IFI35/SP110/PLSCR1/NLRC5/IL15RA/STAT1/HLA-DRB1/SAMHD1/IRF7/SAMD9L/TRIM21/ICAM1/IL15/IFITM3/NMI/HELZ2/CASP4/ZNFX1/HERC6/SECTM1/IRF1/TRIM14/MVP/HLA-B/BPGM/EIF2AK2/LAP3/B2M/PNPT1/PML/TDRD7/VAMP5/MYD88/CD74/STAT2/TNFAIP3/HLA-A/VAMP8/PSME2/UPP1/MX1/SSPN/CASP7/OGFR/CXCL10/AUTS2/CASP8/TOR1B/TAPBP/TRAFD1/BANK1/SLC25A28/NAMPT/STAT4/WARS1/IFITM2 |
| 3 | HALLMARK_G2M_CHECKPOINT | HALLMARK_G2M_CHECKPOINT | 197 | -0.743567453282603 | -3.13792520704078 | 9.27457464770145e-40 | 1.54576244128358e-38 | 6.1830497651343e-39 | 1229 | tags=51%, list=9%, signal=47% | ATF5/SFPQ/CDK4/STAG1/TRA2B/FANCC/CKS2/POLQ/FOXN3/SAP30/CASP8AP2/MAP3K20/HMGN2/RBM14/SRSF1/SMC1A/KPNA2/LBR/CKS1B/RACGAP1/BUB1/H2AZ1/CDC25B/INCENP/TFDP1/HNRNPD/DTYMK/BRCA2/POLE/CHAF1A/SRSF2/MCM3/UBE2S/STIL/CCNF/NASP/DBF4/KIF22/KIF20B/SMC4/MAD2L1/EZH2/H2AZ2/TTK/STMN1/MCM5/POLA2/CDC6/E2F1/MCM2/AURKA/DDX39A/RAD54L/NSD2/TACC3/CCNB2/H2AX/MCM6/CENPE/SMC2/TMPO/CDC25A/BARD1/CENPA/NEK2/KIF15/HMMR/CDK1/UBE2C/RBL1/PRC1/NDC80/CHEK1/CDKN3/KIF11/TPX2/CDKN2C/EXO1/ESPL1/CDC45/GINS2/CDC20/FBXO5/BIRC5/KIF4A/MKI67/KIF2C/PLK1/PBK/CENPF/TOP2A/CCNA2/E2F2/LMNB1/KIF23/TROAP/PTTG1/MYBL2/KNL1/AURKB |
| 4 | HALLMARK_INTERFERON_ALPHA_RESPONSE | HALLMARK_INTERFERON_ALPHA_RESPONSE | 91 | 0.882357420451324 | 2.80515998040605 | 3.14882510753731e-32 | 3.93603138442164e-31 | 1.57441255376865e-31 | 1085 | tags=77%, list=8%, signal=72% | RSAD2/IFIT3/IFIT2/SAMD9/IFI27/CMPK2/OASL/IFI44L/OAS1/BATF2/DDX60/RTP4/ISG15/IFI44/DHX58/IFITM1/IFIH1/TMEM140/PARP9/BST2/UBE2L6/UBA7/ISG20/PSMB9/PARP12/PSMB8/GBP4/USP18/LGALS3BP/PARP14/IRF9/TAP1/IFI35/SP110/PLSCR1/CSF1/LAMP3/IRF7/SAMD9L/TRIM21/TRIM5/IL15/IFITM3/NMI/HELZ2/HERC6/IRF1/TRIM14/EIF2AK2/LAP3/GBP2/B2M/PNPT1/TDRD7/HLA-C/CD74/STAT2/TENT5A/PSME2/MX1/SELL/OGFR/CXCL10/CASP8/CNP/TRAFD1/SLC25A28/WARS1/IFITM2/GMPR |
| 5 | HALLMARK_MYC_TARGETS_V1 | HALLMARK_MYC_TARGETS_V1 | 199 | -0.651790896989032 | -2.75739072143028 | 1.52633201976361e-24 | 1.52633201976361e-23 | 6.10532807905444e-24 | 2893 | tags=53%, list=20%, signal=43% | CYC1/NME1/PABPC1/EIF3D/HNRNPU/GSPT1/NCBP1/CBX3/CCT3/NCBP2/SSBP1/PSMD8/TCP1/PA2G4/PTGES3/YWHAE/CCT4/SERBP1/ILF2/PSMA2/SMARCC1/PWP1/PPIA/GOT2/HNRNPR/RPL34/TXNL4A/HDDC2/XPO1/EIF1AX/VBP1/STARD7/RAN/SNRPD3/RUVBL2/XRCC6/TARDBP/RPL22/SLC25A3/LSM2/ODC1/BUB3/HPRT1/VDAC3/POLD2/CTPS1/CDK2/SNRPD2/SNRPG/SNRPD1/CDK4/IMPDH2/SET/TRIM28/RANBP1/TRA2B/RPL14/RPLP0/MRPL9/PRPS2/SNRPA1/NPM1/RPS10/RPL6/RPL18/HDGF/YWHAQ/RPS5/HNRNPA2B1/RACK1/SRSF1/KPNA2/RPS6/SF3B3/CAD/H2AZ1/RPS2/EEF1B2/NAP1L1/SRSF7/SRSF3/RPS3/TFDP1/HNRNPD/MCM4/DUT/RFC4/HNRNPA1/SRSF2/USP1/RRM1/DEK/MAD2L1/PCNA/SNRPA/PHB2/MCM5/MCM2/FBL/MCM6/MCM7/CDC45/CDC20/TYMS/CCNA2 |
| 6 | HALLMARK_INFLAMMATORY_RESPONSE | HALLMARK_INFLAMMATORY_RESPONSE | 116 | 0.746586465382548 | 2.44187415414222 | 5.95830659973987e-17 | 4.96525549978322e-16 | 1.98610219991329e-16 | 1685 | tags=44%, list=12%, signal=39% | SERPINE1/RTP4/IFITM1/TNFSF10/CCL22/TNFSF15/CCL5/BST2/ADM/EREG/IL15RA/RGS16/CSF1/LAMP3/IRF7/ICAM1/ITGA5/INHBA/IL15/P2RY2/NMI/BDKRB1/IRF1/SLC31A2/RASGRP1/IL18/EIF2AK2/ABCA1/BEST1/HBEGF/PTAFR/CD82/AHR/P2RX4/SPHK1/CD70/BTG2/SELL/CXCL10/TAPBP/NAMPT/EDN1/ADGRE1/SLC1A2/CDKN1A/SLC7A2/MXD1/CCRL2/CCR7/FZD5/RNF144B |
| 7 | HALLMARK_TNFA_SIGNALING_VIA_NFKB | HALLMARK_TNFA_SIGNALING_VIA_NFKB | 162 | 0.657660846206331 | 2.24218393134382 | 4.60340940156814e-13 | 3.28814957254867e-12 | 1.31525982901947e-12 | 2102 | tags=48%, list=15%, signal=41% | IFIT2/SERPINE1/TNFAIP2/DDX58/IFIH1/CCL5/BIRC3/TAP1/IL15RA/CLCF1/PLAU/CSF1/ICAM1/INHBA/LAMB3/IRF1/RELB/SAT1/IL18/RCAN1/FOSB/VEGFA/ABCA1/JAG1/HBEGF/PHLDA2/B4GALT1/SLC16A6/TNFAIP3/DUSP1/BHLHE40/FOS/AREG/DUSP4/TNFAIP8/SPHK1/SMAD3/BTG2/SQSTM1/CXCL10/NR4A3/FJX1/RHOB/GADD45A/ZFP36/ATF3/NAMPT/PHLDA1/NINJ1/EDN1/HES1/BCL6/CDKN1A/SLC2A6/BCL3/KDM6B/MXD1/TRIP10/IER3/PMEPA1/NFKBIE/JUNB/CCRL2/CCL4/MAP2K3/ACKR3/NFKB2/CCND1/LITAF/BMP2/SDC4/TRIB1/LIF/EFNA1/DNAJB4/ZC3H12A/PFKFB3 |
| 8 | HALLMARK_MITOTIC_SPINDLE | HALLMARK_MITOTIC_SPINDLE | 194 | -0.516984600738668 | -2.17786024416096 | 1.77375353915008e-11 | 1.1085959619688e-10 | 4.43438384787521e-11 | 1599 | tags=29%, list=11%, signal=26% | ARHGAP5/TUBGCP3/ALMS1/UXT/CEP131/ECT2/TBCD/CEP57/PREX1/NUMA1/MYH10/CEP192/KPTN/ARHGAP10/CEP250/CENPJ/CDK5RAP2/SMC3/SMC1A/RACGAP1/BUB1/TIAM1/INCENP/FARP1/PCNT/BRCA2/SAC3D1/KNTC1/KIF22/KIF20B/SMC4/PLEKHG2/TTK/ANLN/AURKA/CCNB2/CENPE/NEK2/KIF15/CDK1/PRC1/NDC80/KIF11/TPX2/DLGAP5/ESPL1/PIF1/FBXO5/BIRC5/KIF4A/KIF2C/PLK1/CENPF/TOP2A/LMNB1/KIF23 |
| 9 | HALLMARK_SPERMATOGENESIS | HALLMARK_SPERMATOGENESIS | 70 | -0.663992457014828 | -2.36183724811574 | 8.01167091066527e-10 | 4.45092828370293e-9 | 1.78037131348117e-9 | 2154 | tags=43%, list=15%, signal=37% | RPL39L/IFT88/PRKAR2A/NPHP1/MAST2/TSN/VDAC3/PSMG1/MAP7/OAZ3/TOPBP1/ARL4A/BUB1/RFC4/THEG/DBF4/EZH2/HSPA2/TTK/AURKA/CCNB2/CNIH2/NEK2/CDK1/NCAPH/CDKN3/YBX2/DMC1/KIF2C/ACE |
| 10 | HALLMARK_COMPLEMENT | HALLMARK_COMPLEMENT | 133 | 0.626850640738171 | 2.08904430779541 | 3.82015450624892e-9 | 1.91007725312446e-8 | 7.64030901249784e-9 | 1972 | tags=39%, list=14%, signal=34% | SERPINE1/CCL5/MMP13/PSMB9/S100A9/PLSCR1/IRF7/CASP4/IRF1/RASGRP1/CP/CLU/LAP3/CTSL/TIMP2/CTSO/CD59/TNFAIP3/CPM/CTSS/DOCK4/TMPRSS6/ERAP2/CASP7/CTSH/CTSD/LGALS3/CTSC/TFPI2/CPQ/CD36/CTSB/LIPA/ADAM9/PDGFB/SERPINA1/LGMN/CASP9/PRKCD/CASP10/GZMB/FDX1/SRC/CASP3/GZMA/SPOCK2/ATOX1/CSRP1/IRF2/CD46/CBLB/PHEX |
Showing 1 to 10 of 50 entries
gsea_df$PAR_ARCvsDMSO %>% datatable(caption="PAR_ARCvsDMSO")
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HALLMARK_INTERFERON_GAMMA_RESPONSE | HALLMARK_INTERFERON_GAMMA_RESPONSE | 159 | 0.638030268128343 | 2.07271409921154 | 2.65288128927941e-10 | 1.32644064463971e-8 | 4.7472612545e-9 | 1859 | tags=32%, list=13%, signal=28% | TNFAIP2/IFI27/HLA-DRB1/OASL/ISG20/ISG15/IFIT1/OAS2/BST2/IRF9/CDKN1A/UPP1/IFIT3/DDX60/OAS3/DDX58/SSPN/TRIM14/CCL5/TNFSF10/PLSCR1/MTHFD2/STAT1/NAMPT/WARS1/B2M/HLA-B/ZNFX1/RSAD2/PNP/LY6E/SP110/SAMHD1/VAMP8/VAMP5/HLA-A/SLC25A28/SECTM1/IFIH1/LGALS3BP/PFKP/IL15RA/FAS/IFITM3/HLA-DMA/MT2A/RBCK1/LAP3/GCH1/HELZ2/PSME2 |
| 2 | HALLMARK_P53_PATHWAY | HALLMARK_P53_PATHWAY | 185 | 0.60606155751223 | 2.00157143665141 | 4.50504284664892e-9 | 1.12626071166223e-7 | 4.03082781015956e-8 | 2369 | tags=41%, list=16%, signal=35% | TM4SF1/NUPR1/PLK3/SLC7A11/TGFA/VDR/CDKN2B/CDKN1A/UPP1/SLC3A2/ADA/DGKA/GADD45A/IER5/EPHA2/NDRG1/IER3/SPHK1/SDC1/DDIT3/STOM/SAT1/HBEGF/OSGIN1/ABAT/RGS16/CTSD/INHBB/TOB1/ATF3/ST14/AEN/KRT17/ZMAT3/APP/SFN/VAMP8/HEXIM1/MDM2/MXD1/CD81/TRIB3/FUCA1/RPS27L/PHLDA3/BAK1/GM2A/ABHD4/PVT1/POLH/HMOX1/RAD51C/DDB2/HSPA4L/ITGB4/FAS/ZNF365/LIF/ANKRA2/TSPYL2/CD82/KLF4/S100A10/MKNK2/STEAP3/PERP/RETSAT/TSC22D1/EPS8L2/PROCR/TM7SF3/PTPRE/BLCAP/NOTCH1/TP63/FDXR |
| 3 | HALLMARK_COAGULATION | HALLMARK_COAGULATION | 73 | 0.704900837465225 | 2.1069318247513 | 1.85141802794598e-7 | 0.00000296586223132408 | 0.00000106146648278967 | 1327 | tags=37%, list=9%, signal=34% | RAPGEF3/CAPN5/THBD/SERPINE1/TIMP3/CLU/CTSH/TFPI2/FN1/DUSP6/PRSS23/ITGA2/APOC1/PDGFB/F12/CTSB/KLF7/CSRP1/TIMP1/PLAU/ANXA1/GSN/ACOX2/CTSV/S100A13/LGMN/LAMP2 |
| 4 | HALLMARK_KRAS_SIGNALING_UP | HALLMARK_KRAS_SIGNALING_UP | 118 | 0.619577052806343 | 1.96000385654841 | 4.64875880065027e-7 | 0.00000296586223132408 | 0.00000106146648278967 | 1048 | tags=25%, list=7%, signal=23% | KCNN4/TSPAN1/HDAC9/MPZL2/EREG/TFPI/EMP1/ALDH1A3/TRIB2/CPE/ITGB2/SEMA3B/NRP1/INHBA/GADD45G/ANGPTL4/DUSP6/PLEK2/HBEGF/ETV5/RGS16/ITGA2/SLPI/BTC/APOD/PLAUR/PLAU/CROT/FUCA1 |
| 5 | HALLMARK_TNFA_SIGNALING_VIA_NFKB | HALLMARK_TNFA_SIGNALING_VIA_NFKB | 162 | 0.585109863960566 | 1.90980344495504 | 4.74537957011853e-7 | 0.00000296586223132408 | 0.00000106146648278967 | 2042 | tags=36%, list=14%, signal=32% | TNFAIP2/JAG1/AREG/SLC16A6/SERPINE1/DUSP4/CDKN1A/B4GALT1/PHLDA2/GADD45A/IER5/INHBA/IER3/SPHK1/DDX58/SAT1/HBEGF/PFKFB3/CCL5/CLCF1/TRIP10/MAP3K8/PHLDA1/RCAN1/LAMB3/EDN1/DUSP1/NAMPT/ATF3/TNFAIP8/VEGFA/DUSP5/PLAUR/SDC4/PLAU/FOSL1/CCND1/MXD1/IL18/GADD45B/SERPINB8/LITAF/MAP2K3/KYNU/IFIH1/SLC2A6/BCL3/ZFP36/IL15RA/JUNB/LIF/GCH1/BTG3/KLF4/SQSTM1/ZC3H12A/IFIT2/BCL6/TSC22D1 |
| 6 | HALLMARK_MTORC1_SIGNALING | HALLMARK_MTORC1_SIGNALING | 189 | 0.571191653193619 | 1.88799237949741 | 3.87790991923396e-7 | 0.00000296586223132408 | 0.00000106146648278967 | 2819 | tags=42%, list=20%, signal=34% | NUPR1/TUBA4A/SLC7A11/STC1/ITGB2/CDKN1A/RRM2/SLC7A5/G6PD/HK2/DDIT3/PSPH/PHGDH/ME1/MTHFD2/SLC1A4/HSPA5/NAMPT/WARS1/ATP5MC1/SDF2L1/CTH/SHMT2/POLR3G/SLC1A5/PNP/CALR/ACSL3/PGM1/PRDX1/TRIB3/ELOVL5/SLC2A1/GOT1/CTSC/MAP2K3/AK4/ELOVL6/MLLT11/EIF2S2/GLRX/LGMN/EPRS1/TXNRD1/DHCR24/HSP90B1/SERPINH1/SLC6A6/IDH1/VLDLR/DAPP1/CXCR4/GSR/SLC9A3R1/GLA/PIK3R3/PGK1/SQSTM1/PSMC4/EDEM1/SCD/RDH11/ERO1A/AURKA/TUBG1/SSR1/FDXR/HSPE1/ACTR3/GPI/NFKBIB/ATP6V1D/ENO1/ATP2A2/RPN1/CANX/M6PR/UCHL5/HSPD1 |
| 7 | HALLMARK_GLYCOLYSIS | HALLMARK_GLYCOLYSIS | 175 | 0.571098583811086 | 1.87324259436125 | 4.26375564741532e-7 | 0.00000296586223132408 | 0.00000106146648278967 | 2216 | tags=37%, list=15%, signal=32% | NT5E/B3GNT3/ISG20/GPR87/QSOX1/STC1/TGFBI/TGFA/CAPN5/B4GALT1/SLC16A3/G6PD/HK2/IER3/SDC1/ANGPTL4/GALE/HS6ST2/ELF3/PYGL/ME1/HSPA5/KDELR3/CTH/RRAGD/P4HA2/SPAG4/B3GAT3/DSC2/VEGFA/B4GALT4/UGP2/PAM/GOT1/SLC25A13/AK4/FUT8/CLDN3/CDK1/PKM/PFKP/GLRX/GPC4/EXT2/SRD5A3/AK3/EGFR/IDH1/VLDLR/RBCK1/CXCR4/HMMR/PGK1/HAX1/CHPF/TALDO1/SOX9/CLDN9/BIK/PSMC4/ERO1A/AURKA/ANKZF1/CENPA/DLD |
| 8 | HALLMARK_ESTROGEN_RESPONSE_EARLY | HALLMARK_ESTROGEN_RESPONSE_EARLY | 197 | 0.563185356031477 | 1.86762883336863 | 4.61313962497054e-7 | 0.00000296586223132408 | 0.00000106146648278967 | 1508 | tags=38%, list=10%, signal=34% | DHRS2/AQP3/DHRS3/ALDH3B1/KCNK15/SCNN1A/CLIC3/CCN5/NAV2/TSKU/FRK/P2RY2/HSPB8/MUC1/AREG/SEMA3B/CA12/PAPSS2/B4GALT1/FHL2/SLC7A5/CLDN7/SULT2B1/SYNGR1/PTGES/TFF1/PLAAT3/ZNF185/RET/BLVRB/PRSS23/LAD1/RASGRP1/SH3BP5/SLC1A1/PODXL/ABAT/SLC7A2/KRT18/ANXA9/ELF3/THSD4/KRT8/RPS6KA2/INHBB/SLC1A4/SEC14L2/IGFBP4/TOB1/ENDOD1/SYT12/CELSR2/WFS1/ABLIM1/SFN/CISH/MICB/PEX11A/CCND1/ELOVL5/CALCR/SLC2A1/RHOBTB3/MED13L/KRT19/TFAP2C/MSMB/FAM102A/RAB17/CANT1/PDLIM3/BAG1/RHOD/KRT15 |
| 9 | HALLMARK_INFLAMMATORY_RESPONSE | HALLMARK_INFLAMMATORY_RESPONSE | 116 | 0.619843162758208 | 1.95962477377395 | 6.47616673947317e-7 | 0.00000359787041081843 | 0.00000128765888387186 | 2274 | tags=37%, list=16%, signal=31% | ADM/EREG/P2RY2/ITGA5/BST2/PTAFR/SERPINE1/CDKN1A/CCL22/INHBA/SPHK1/HBEGF/RASGRP1/SLC7A2/RGS16/CCL5/FFAR2/TNFSF10/SLC1A2/EDN1/NAMPT/SLC31A1/PVR/PLAUR/LY6E/SLC31A2/TIMP1/MXD1/IL18/BEST1/OSMR/IL15RA/LIF/CD82/BDKRB1/TNFSF15/GCH1/FZD5/CD55/ICAM1/NMI/EIF2AK2/PTPRE |
| 10 | HALLMARK_INTERFERON_ALPHA_RESPONSE | HALLMARK_INTERFERON_ALPHA_RESPONSE | 91 | 0.656262653129685 | 2.01359096759233 | 8.1391779859818e-7 | 0.0000040695889929909 | 0.00000145648448170201 | 2573 | tags=44%, list=18%, signal=36% | OAS1/IFI27/OASL/ISG20/ISG15/BST2/IRF9/TENT5A/PARP9/IFIT3/DDX60/TRIM14/PLSCR1/WARS1/B2M/GBP2/RSAD2/LY6E/SP110/SLC25A28/TMEM140/IFIH1/LGALS3BP/SAMD9/TRIM5/IFITM3/NCOA7/LAP3/HELZ2/PSME2/IFIT2/IRF2/PROCR/NMI/EIF2AK2/CMPK2/IRF7/UBE2L6/CD47/TRIM21 |
Showing 1 to 10 of 50 entries
gsea_df$PAR_LY.ARCvsDMSO %>% datatable(caption = "PAR_LY.ARCvsDMSO")
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HALLMARK_E2F_TARGETS | HALLMARK_E2F_TARGETS | 200 | -0.827252327210745 | -3.36994897605431 | 1.45893762232686e-56 | 7.29468811163428e-55 | 3.22502000724884e-55 | 1697 | tags=69%, list=12%, signal=62% | NUDT21/SHMT1/CSE1L/LUC7L3/UBR7/CDKN1B/CTPS1/ZW10/HMGB3/CNOT9/TP53/NAP1L1/XPO1/MRE11/MSH2/PHF5A/RAD51C/POLD1/NUP107/GINS4/TRA2B/DCLRE1B/SMC6/SMC3/CDK4/LYAR/POLD3/PRKDC/RPA3/SMC1A/SLBP/CDC25B/TUBB/SNRPB/DNMT1/RANBP1/UNG/UBE2T/GINS3/TUBG1/SRSF2/SUV39H1/CCP110/EXOSC8/SRSF1/RFC3/ORC6/RNASEH2A/USP1/RFC2/KPNA2/LBR/HNRNPD/TIPIN/PLK4/SSRP1/MCM3/MCM4/ANP32E/H2AZ1/DUT/NASP/EZH2/POLE/DEK/KIF22/NCAPD2/CBX5/TIMELESS/CKS2/STMN1/SMC4/RACGAP1/MCM6/MMS22L/GINS1/PSIP1/E2F8/LIG1/CKS1B/HELLS/DDX39A/MCM2/POLA2/PCNA/AURKA/MCM5/TMPO/BRCA1/CDC25A/CHEK1/MAD2L1/BARD1/H2AX/MCM7/ATAD2/CDCA8/RAD51AP1/TRIP13/CDKN3/BRCA2/MELK/CCNB2/SPC24/CDKN2C/SPAG5/BUB1B/DSCC1/PSMC3IP/CENPM/ASF1B/CENPE/TACC3/ESPL1/LMNB1/TCF19/RRM2/CIT/HMMR/TK1/HMGB2/CDCA3/DLGAP5/TOP2A/DEPDC1/MXD3/KIF18B/MKI67/CDK1/CDC20/PTTG1/BIRC5/KIF4A/SPC25/MYBL2/PLK1/KIF2C/AURKB |
| 2 | HALLMARK_G2M_CHECKPOINT | HALLMARK_G2M_CHECKPOINT | 197 | -0.806189846619238 | -3.27902545979478 | 6.86826763437273e-49 | 1.71706690859318e-47 | 7.59124317483302e-48 | 1029 | tags=52%, list=7%, signal=49% | MAP3K20/CDC7/HMGN2/CDK4/AMD1/MT2A/SMC1A/SNRPD1/SAP30/CASP8AP2/TFDP1/CDC25B/SRSF2/SUV39H1/TRAIP/RBM14/SRSF1/ORC6/KPNA2/LBR/HNRNPD/PLK4/ATF5/MCM3/H2AZ1/DTYMK/NASP/EZH2/POLE/DBF4/STIL/CHAF1A/KIF22/CCNF/INCENP/CKS2/NSD2/STMN1/SMC4/H2AZ2/RACGAP1/MCM6/E2F1/POLQ/CKS1B/BUB1/DDX39A/MCM2/POLA2/AURKA/KIF20B/MCM5/TMPO/CDC25A/CHEK1/KIF23/SMC2/MAD2L1/BARD1/H2AX/GINS2/RBL1/FBXO5/CDKN3/BRCA2/CCNB2/CDKN2C/KIF11/CENPE/TPX2/CDC6/TTK/TACC3/ESPL1/NEK2/EXO1/PRC1/LMNB1/UBE2C/CDC45/CENPA/NDC80/CENPF/PBK/HMMR/TOP2A/MKI67/CDK1/E2F2/CDC20/KNL1/PTTG1/TROAP/CCNA2/BIRC5/KIF4A/MYBL2/PLK1/KIF15/RAD54L/KIF2C/AURKB |
| 3 | HALLMARK_INTERFERON_GAMMA_RESPONSE | HALLMARK_INTERFERON_GAMMA_RESPONSE | 159 | 0.794913716995598 | 2.86675159330622 | 2.61601913624887e-34 | 4.36003189374811e-33 | 1.92759304776232e-33 | 1229 | tags=49%, list=9%, signal=45% | RSAD2/IFIT1/IFIT3/OAS2/IFI27/IFIT2/DDX60/TNFAIP2/ISG15/MX2/CMPK2/IFI44L/OASL/XAF1/DDX58/IFIH1/BATF2/TNFSF10/DHX58/IFI44/BST2/UBE2L6/RTP4/ISG20/IL15/OAS3/CCL5/PARP14/LGALS3BP/IRF9/PARP12/SP110/APOL6/STAT1/IL15RA/NLRC5/IRF7/HLA-DRB1/PLSCR1/MVP/TAP1/BPGM/ZNFX1/USP18/HELZ2/VAMP5/IFI35/TRIM14/HERC6/IRF1/IFITM3/CASP4/PSMB9/TRIM21/ICAM1/NMI/SAMHD1/EIF2AK2/AUTS2/PSMB8/SECTM1/HLA-B/VAMP8/GBP4/LAP3/UPP1/SSPN/STAT2/MYD88/CDKN1A/B2M/CASP7/PML/STAT4/CD74/HLA-A/TDRD7/PNPT1 |
| 4 | HALLMARK_INTERFERON_ALPHA_RESPONSE | HALLMARK_INTERFERON_ALPHA_RESPONSE | 91 | 0.836072944238735 | 2.80153481359722 | 2.08445797958037e-25 | 2.60557247447546e-24 | 1.15193730450494e-24 | 784 | tags=56%, list=5%, signal=53% | RSAD2/IFIT3/SAMD9/IFI27/IFIT2/DDX60/OAS1/ISG15/CMPK2/TMEM140/IFI44L/OASL/PARP9/IFIH1/IFITM1/BATF2/DHX58/IFI44/BST2/UBE2L6/RTP4/ISG20/IL15/PARP14/LGALS3BP/IRF9/PARP12/SP110/IRF7/PLSCR1/TAP1/TRIM5/USP18/HELZ2/IFI35/TRIM14/HERC6/CSF1/GBP2/IRF1/IFITM3/PSMB9/TRIM21/HLA-C/NMI/EIF2AK2/PSMB8/TENT5A/UBA7/GBP4/LAP3 |
| 5 | HALLMARK_MYC_TARGETS_V1 | HALLMARK_MYC_TARGETS_V1 | 199 | -0.65142823486878 | -2.6500221071573 | 4.99294495450904e-21 | 4.99294495450904e-20 | 2.2074072430461e-20 | 2779 | tags=51%, list=19%, signal=42% | UBA2/ABCE1/LSM7/RPL34/XRCC6/RUVBL2/SRPK1/SMARCC1/DHX15/PPIA/CCT7/EIF4A1/CCT4/HDAC2/HNRNPU/PWP1/YWHAE/RAN/CCT5/SSBP1/GOT2/RPLP0/VBP1/PA2G4/PTGES3/POLD2/SERBP1/CBX3/BUB3/SNRPD3/NCBP1/PSMD8/EEF1B2/RPL14/SNRPD2/HNRNPR/RPS6/IMPDH2/HDDC2/ERH/LSM2/RPS2/TARDBP/SNRPG/CTPS1/SET/PSMD3/PSMA2/RPL18/NAP1L1/XPO1/RPS5/EIF1AX/ILF2/RACK1/PRPS2/TRA2B/YWHAQ/MRPL9/SNRPA1/CAD/CDK4/VDAC3/RPS10/CDK2/SNRPD1/NPM1/TFDP1/HPRT1/TRIM28/HDGF/RANBP1/RPS3/SRSF2/SF3B3/SRSF1/HNRNPA1/USP1/HNRNPA2B1/KPNA2/SRSF3/HNRNPD/SRSF7/MCM4/H2AZ1/DUT/RRM1/PHB2/DEK/RFC4/SNRPA/FBL/MCM6/MCM2/PCNA/MCM5/MAD2L1/MCM7/CDC45/TYMS/CDC20/CCNA2 |
| 6 | HALLMARK_MITOTIC_SPINDLE | HALLMARK_MITOTIC_SPINDLE | 194 | -0.614763859892233 | -2.49367491659971 | 1.89680244967899e-16 | 1.58066870806582e-15 | 6.98821955144891e-16 | 1207 | tags=26%, list=8%, signal=24% | PREX1/SAC3D1/NUMA1/TBCD/SMC3/EPB41L2/ALMS1/ARHGAP10/KPTN/TIAM1/CDC42EP4/FARP1/CDK5RAP2/SMC1A/PCNT/ECT2/CNTRL/CEP250/SASS6/PLEKHG2/KIF22/INCENP/SMC4/RACGAP1/BUB1/AURKA/KIF20B/KIF23/FBXO5/BRCA2/CCNB2/KIF11/CENPE/TPX2/TTK/ESPL1/NEK2/PRC1/LMNB1/NDC80/CENPF/DLGAP5/TOP2A/CDK1/ANLN/PIF1/BIRC5/KIF4A/PLK1/KIF15/KIF2C |
| 7 | HALLMARK_INFLAMMATORY_RESPONSE | HALLMARK_INFLAMMATORY_RESPONSE | 116 | 0.714488335049173 | 2.46301303671195 | 1.89146415595212e-15 | 1.35104582568009e-14 | 5.97304470300669e-15 | 1567 | tags=42%, list=11%, signal=38% | SERPINE1/IFITM1/TNFSF15/BDKRB1/RGS16/TNFSF10/ADM/CCL22/BST2/RTP4/EREG/IL15/CCL5/IL18/IL15RA/P2RY2/BEST1/IRF7/SLC31A2/ITGA5/INHBA/CSF1/RASGRP1/IRF1/HBEGF/ABCA1/SLC7A2/ICAM1/P2RX4/NMI/EIF2AK2/CD82/BTG2/PTAFR/SPHK1/EDN1/CDKN1A/AHR/SLC1A2/SLC4A4/TNFSF9/LIF/MXD1/FZD5/TAPBP/FFAR2/CD14/NAMPT/SELENOS |
| 8 | HALLMARK_SPERMATOGENESIS | HALLMARK_SPERMATOGENESIS | 70 | -0.730213676824621 | -2.5506468264665 | 5.46408577330064e-12 | 3.4150536083129e-11 | 1.50981317420149e-11 | 1163 | tags=34%, list=8%, signal=32% | RPL39L/PSMG1/TOPBP1/ARL4A/VDAC3/MAP7/HSPA2/ACE/EZH2/DBF4/RFC4/BUB1/THEG/AURKA/DMC1/CDKN3/CCNB2/CNIH2/YBX2/TTK/NEK2/NCAPH/CDK1/KIF2C |
| 9 | HALLMARK_TNFA_SIGNALING_VIA_NFKB | HALLMARK_TNFA_SIGNALING_VIA_NFKB | 162 | 0.596293991265112 | 2.15913951206473 | 8.5563741462112e-11 | 4.75354119233956e-10 | 2.10156557977117e-10 | 2016 | tags=41%, list=14%, signal=36% | IFIT2/SERPINE1/TNFAIP2/DDX58/IFIH1/CCL5/CLCF1/SAT1/IL18/IL15RA/LAMB3/BIRC3/TAP1/PLAU/RCAN1/INHBA/CSF1/RELB/IRF1/SLC16A6/HBEGF/ABCA1/NR4A3/JAG1/ICAM1/VEGFA/B4GALT1/PHLDA2/GADD45A/DUSP1/BTG2/DUSP4/SQSTM1/KLF2/SPHK1/FOSB/EDN1/CDKN1A/PHLDA1/RHOB/SMAD3/ZC3H12A/TNFSF9/FJX1/ZFP36/LIF/CCND1/AREG/MXD1/NINJ1/SLC2A6/BHLHE40/LITAF/NAMPT/IL6ST/PMEPA1/FOS/KDM6B/ACKR3/IER3/TNFAIP8/YRDC/TRIB1/HES1/BCL6/PFKFB3/NFKBIE |
| 10 | HALLMARK_KRAS_SIGNALING_UP | HALLMARK_KRAS_SIGNALING_UP | 118 | 0.565914099907497 | 1.96066886319018 | 0.00000101043566844617 | 0.00000505217834223087 | 0.00000223359463551259 | 2076 | tags=36%, list=14%, signal=31% | TSPAN1/ALDH1A3/RGS16/ST6GAL1/TFPI/EREG/KCNN4/CPE/GLRX/MPZL2/SEMA3B/TRIB2/SOX9/BIRC3/ITGB2/BPGM/GPNMB/PLAU/APOD/INHBA/HBEGF/CXCR4/FUCA1/GADD45G/PSMB8/NRP1/MMP11/ITGA2/ANGPTL4/NAP1L2/LAPTM5/LIF/GYPC/CCND2/CROT/ENG/CTSS/HDAC9/YRDC/TRIB1/BTC/CAB39L |
Showing 1 to 10 of 50 entries
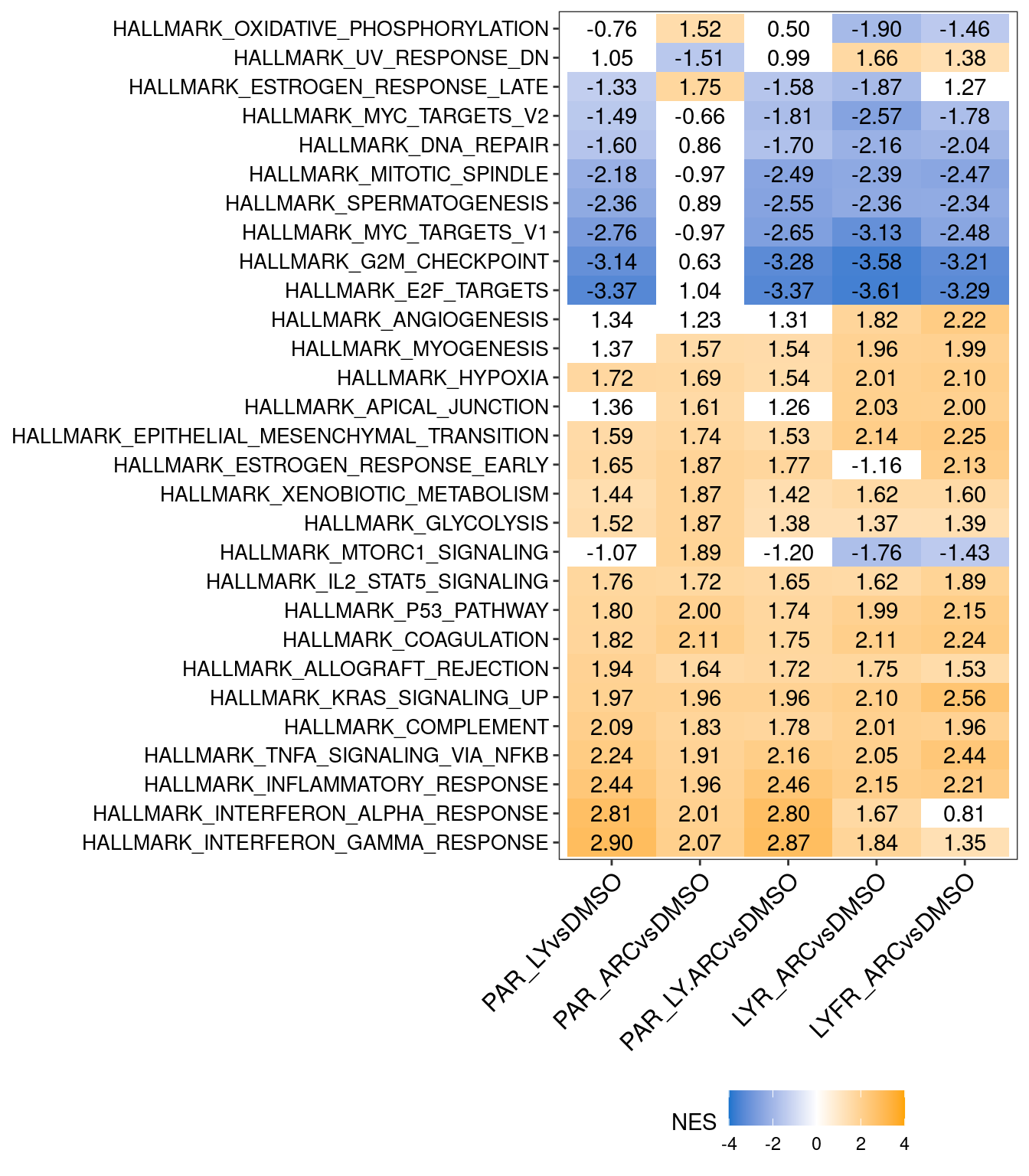
## Warning: Removed 22 rows containing missing values (`geom_point()`).
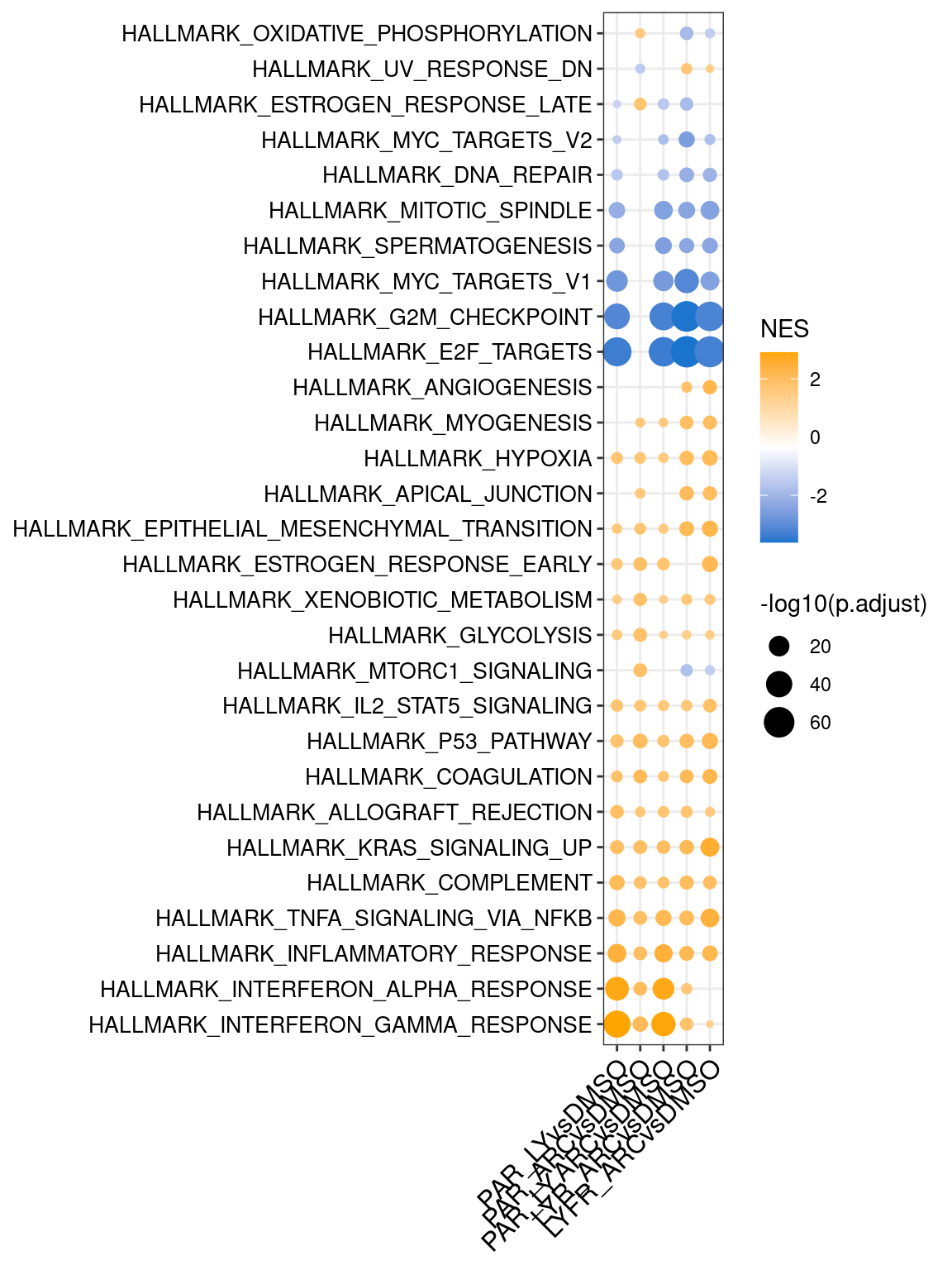
barcode plots
library(clusterProfiler)
library(enrichplot)
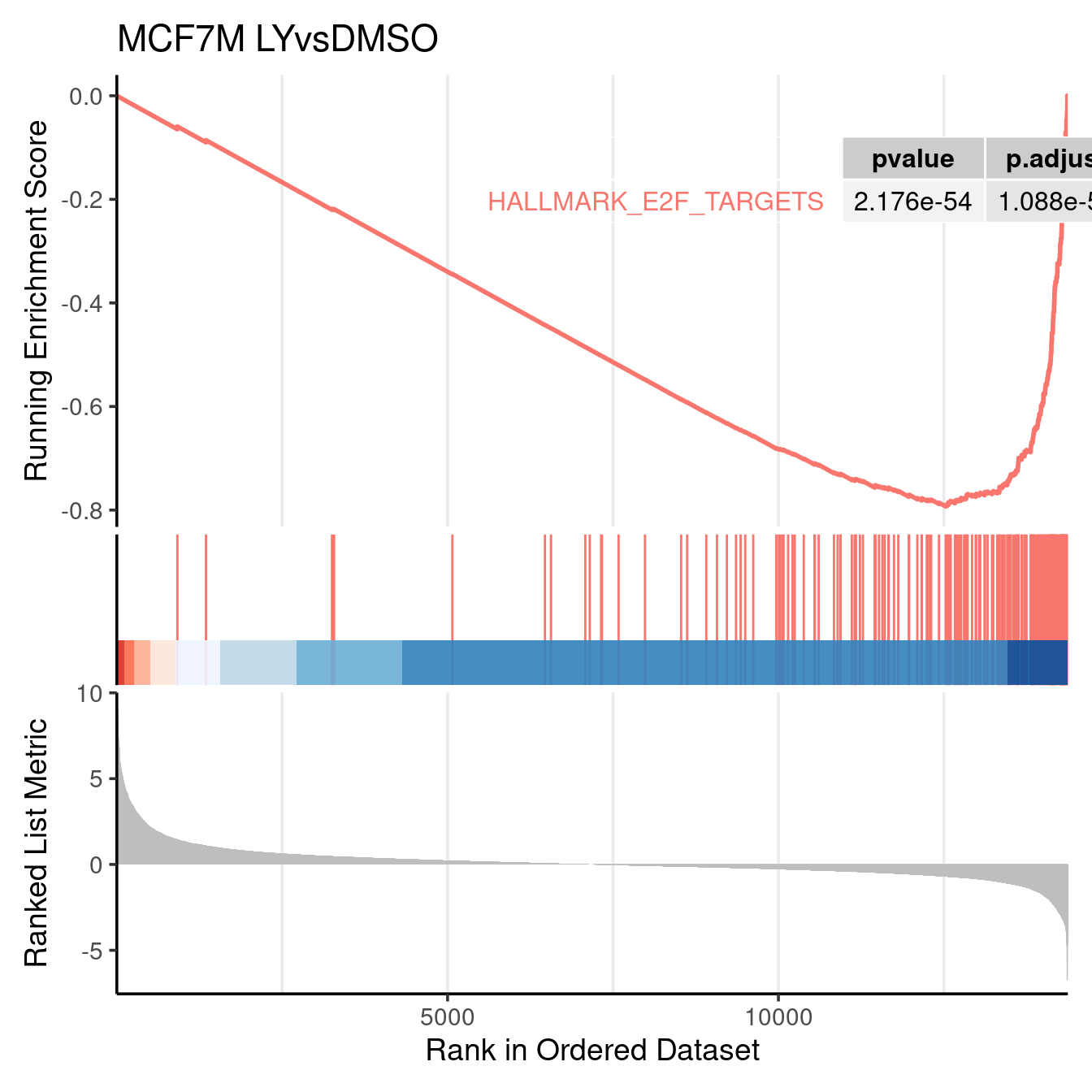
gseaplot2(gsea_output$PAR_LYvsDMSO, geneSetID = c("HALLMARK_E2F_TARGETS"), title = "MCF7M LYvsDMSO", color = c("#E495A5", "#86B875"), pvalue_table = TRUE, base_size = 14)
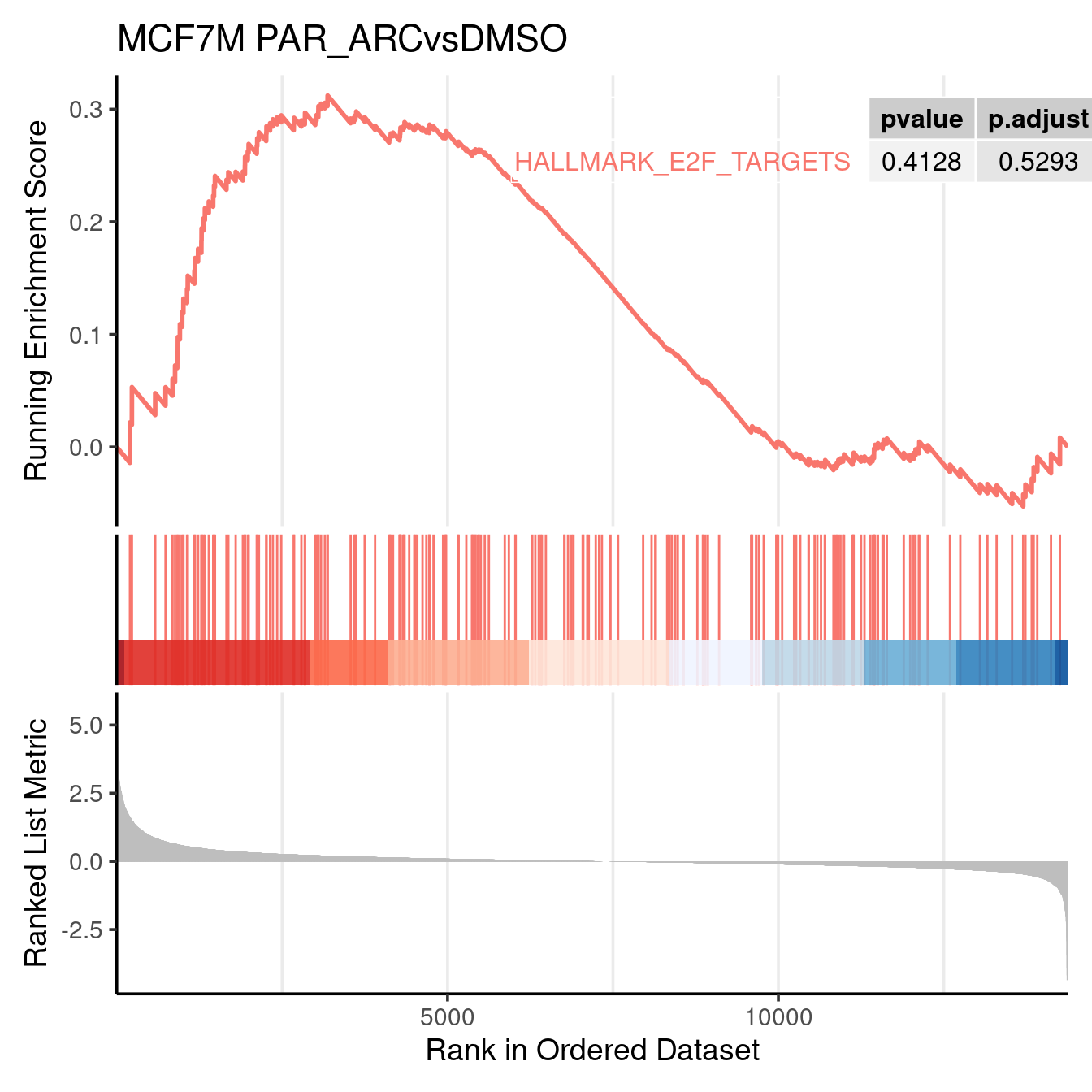
gseaplot2(gsea_output$PAR_ARCvsDMSO, geneSetID = c("HALLMARK_E2F_TARGETS"), title = "MCF7M PAR_ARCvsDMSO", color = c("#E495A5", "#86B875"), pvalue_table = TRUE, base_size = 14)
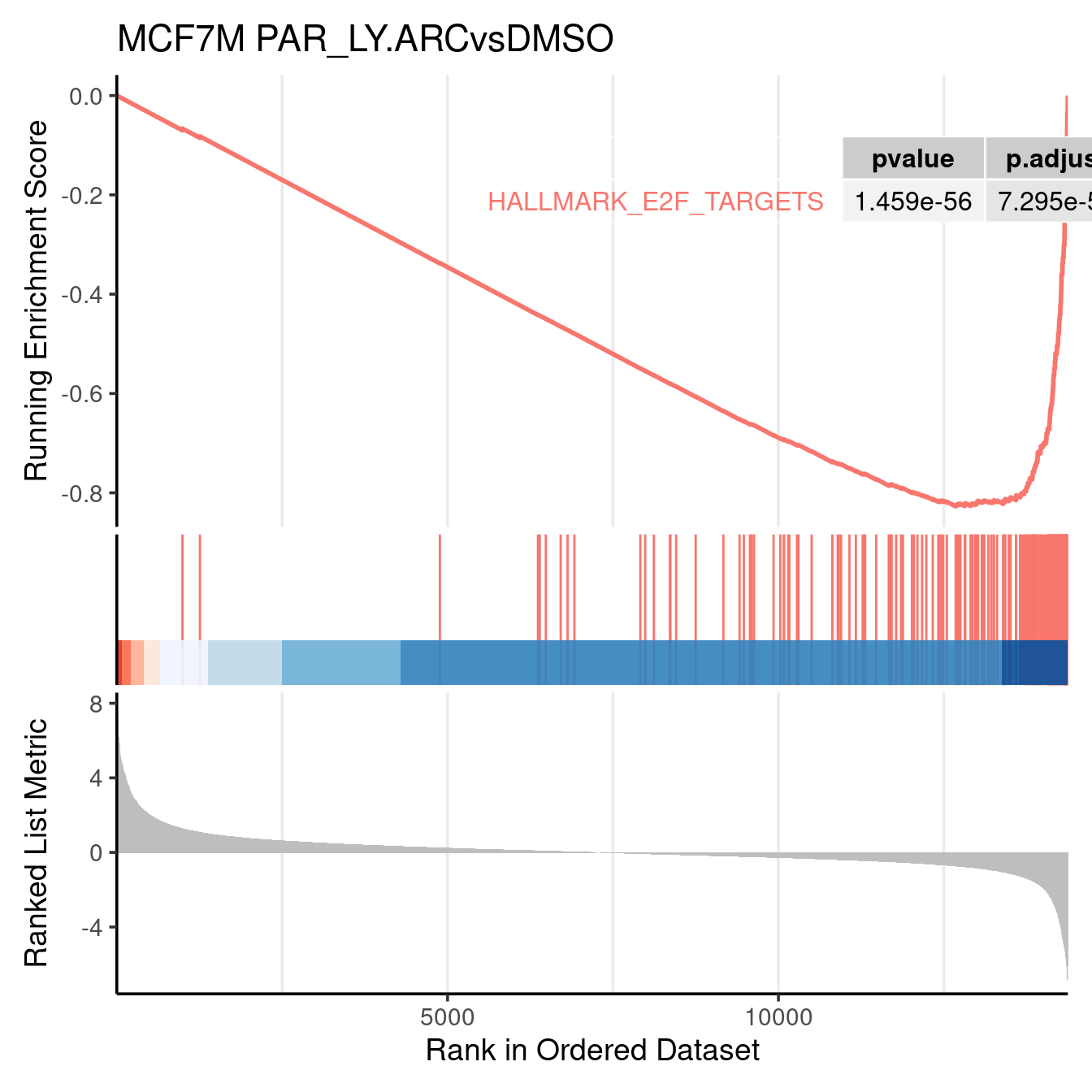
gseaplot2(gsea_output$PAR_LY.ARCvsDMSO, geneSetID = c("HALLMARK_E2F_TARGETS"), title = "MCF7M PAR_LY.ARCvsDMSO", color = c("#E495A5", "#86B875"), pvalue_table = TRUE, base_size = 14)